New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
BE.1.1.1 sublineage with Orf1b:Y264H and S:N460K ( 69 sequences ) emerged in Nigeria ( 14 seqs ) #993
Comments
|
I concur that early attention is in place here. The affinity impact of S:N460K is huge, see https://www.biorxiv.org/content/10.1101/2022.08.07.503115v1.full and https://www.biorxiv.org/content/10.1101/2022.07.18.500332v2.full |
|
It's possible part of the reason BA.2.75 has been able to pick up immune-evasive mutations so much more quickly than BA.5 is because of its higher ACE2 affinity, mostly due to S:N460K. If N460K has a similar effect in BE.1.1.1, it could allow it to acquire still more mutations to evade vaccine- and infection-induced immunity, a factor which will only increase in importance as more and more people are infected with BA.4/5 and/or vaccinated with BA.1- or BA.5-based vaccines. Given that this one is already widespread geographically—possibly far more than we can currently tell—I think it bears close monitoring and probably a rapid designation if more sequences continue to appear in the next week or two. |
This comment was marked as resolved.
This comment was marked as resolved.
|
I'm not very good at using this, so I don't know the meaning of this corresponding time. |
|
@Mydtlwn If you mean the early first date - this is just a normal date error. Happens all the time - probably would be best to scrape that "first date" column from cov-lineages website for that reason or make it more robust (e.g. date of 1% quantile) |
There was ORF1b:K82R in the sibling lineage of Ay.103 that was only partially designated as AY.41 (I opened an issue back months ago) that opened breaches in eastern (balcanic) Europe |
|
New sequence from Georgia state - US collected 8/08/22 : EPI_ISL_14675347 with an additional Orf9b:R13L, Orf1a:A2856V |
|
New sequence from Germany : EPI_ISL_14697838 (this last has one more spike mutation: S:I666V) |
|
3 more sequences from Nigeria (where it likely emerged) EPI_ISL_14711977 all collected on 10th of July. I suggest flagging this sublineage to health authorities , it could be really a step beyond BA.5. cc @corneliusroemer @chrisruis @InfrPopGen EDITED List of sequences from Nigeria: |
|
New Usher tree for the new sequences from Nigeria ( @AngieHinrichs ) confirms at least 14 out of 16 sequences are part of this new sublineage: New things on the table: the two sequence that Usher doesnt place in the S:N460K cluster, one (hCoV-19/Nigeria/NCDC-NR-GL-005829/2022|EPI_ISL_14711966|2022-07-10) is clearly linked and could be considered basal to this new lineage together with a sequence from Usa (USA/DC-Curative-451803/2022|EPI_ISL_14464964|2022-08-06) that has too X aminoacid in the region spanning Spike residues 444-460 so it could be instead part of it. ((while the other one (hCoV-19/Nigeria/NCDC-NR-GL-005827/2022|EPI_ISL_14711964|2022-07-10) is placed in a messy BA.5.5/Ba.5.3 part of the tree: https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/subtreeAuspice2_genome_3f237_d48520.json?branchLabel=aa%20mutations&c=pango_lineage_usher&label=nuc%20mutations:C11750T)) |
|
If we get a few more this should be designated as there's quite some literature on 460K if I'm not mistaken |
|
@corneliusroemer two more sequences from Texas uploaded today and collected 7/08 and 21/08: EPI_ISL_14752457 |
|
Oh bad news EPI_ISL_14752457 the recent Texas sequence got S:346T @corneliusroemer |
|
New sequence new country: Belgium EPI_ISL_14773615 collected on 24/08/2022 |
|
One more sequence from Oregon, Usa: EPI_ISL_14777488 |
|
@FedeGueli feel free to propose the most "promising" lineages in your view, the 346T sound interesting, definitely something to monitor and designate once it gets above 5 or maybe 10. |
|
4 more sequences: Denmark, Rhode Island , Georgia and Canada (intl Airport surveillance from unknown country). I noticed that the belgian one i highlighted above beyond S:346T has a reversion at 484 to E that i missed before. Ok @corneliusroemer i ll wait a bit to have a more clear picture and then i will propose sublineages of it. |
|
4 new seqs from Denmark, Uk and 2 from Germany one of them with S:R346T. Later i ll.propose the S:346T sublineage. |
|
I'll designate the S:R346T straightaway, seeing how fast BQ.1 grows and that the 5 S:R346T are from 5 countries and very recent. |
|
Here's an Usher tree with all sequences (some are duplicated because I uploaded all 66 that the GISAID query found, and they were partially in the tree already): |
…ed designation First mentioned by @FedeGueli in issue #993
|
I've added BQ.1.1 on the ORF1b:N1191S branch defined by S:R346T in commit 44c66bc Thanks @FedeGueli |
|
Thanks @corneliusroemer great work! |
|
Gisaid query for BQ.1.1:Gisaid: Spike_N460K,Spike_K444T,Spike_R346T,Nsp13_N268S |
|
Already 7 BQ.1.1 (two newly uploaded and one of them sampled in Japan coming from Nigeria as noted by @ryhisner .) beyond these two there were 9 BQ.1 uploaded 8 from Usa and 1 from Greece . |
|
There are now 12 BQ.1.1 already |
|
11 BQ.1.1 uploaded yesterday mostly England, and 2 new countries Belgium and Italy. @corneliusroemer @thomaspeacock it seems really fast. |
|
2 danish BQ.1 sequences with a couple of spike mutations: Spike_R190T,Spike_N556K. @ryhisner apotteda couple of BQ.1 with S:T444M T > M that is to be monitored. I suggest to use this modified query: NSP12_Y273H,Spike_K444,Spike_N460K |
|
one more sequence of BQ.1 with S:_R190T but without S:N556K. Total counts as today: |
|
8 BQ.1.1 from.Uk uploaded today all with S:L822F mutation. |
|
From #931:
|
|
I realized this just today by looking at Venn diagram by @RajLABN https://twitter.com/RajlabN/status/1572480532478320642/photo/1 |
|
We don't know if it is relevant of note, but we have noticed that for: |
|
@carlottaolivero thx! great catch! i did a rapid query on covspectrum i found that T22942A is in BQ.1* , XAW and some of the BA.5.2 +460K we saw in the last times. |
|
FYI @corneliusroemer In Germany's RKI DESH DATA are now (since 2022-08-15 / KW33):
|
|
Outbreak Info and Cov-lineages list BQ.1.1 first sequence as showing up in October 15... 2020. Isn't it a mistake? Where does it come from... Could it be from faulty metadata on gisaid? Upon inspecting GISAID, it even flags the "2020" sequence as an outlier, as its collection date is earlier than the clade name submission. Shouldn't outbreak info and cov-lineages filter such sequences out? Thank you for clarifying. |
Most likely yes. If you have time to contact the submitters, sometimes they are responsive and fix the record, sometimes not. Another thing that can happen is that they pull an old sample out of the freezer (so the collection date is true) -- but then contamination happens during sequencing. |
|
I'll try to contact them and I'll let you know if anything changes.
|










Detected a new potential sublineage of interest of recently designated BE.1.1.1.
Defining mutations : Orf1b:Y264H and S:N460K
7 sequences Usa Uk Japan
The one from Japan (EPI_ISL_14632425 )
was intercepted by airport surveillance and has travel history from Egypt via Italy and Israel to Japan and it is heavily mutated with further spike mutations: S:261D*,S:494P and S:603S.
(* as noted by @ryhisner elsewhere S:G261D is or highly homoplasic mutation or pipeline artifact , it was present in a lot of samples from Japanese airport surveillance yesterday maybe Ryan could explain that better than me.)
This could suggest circulation and evolution in an undersampled area of Africa likely , due to the fact that the likely origin of BE.1.1.1 lineage is Nigeria)
Gisaid query: NSP12_Y273H,Spike_K444T,Spike_N460K finds 6 out of 7
CovSpectrum: 4 out of 7 Overview
tree: https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/subtreeAuspice1_genome_3ba8a_876050.json?c=pango_lineage_usher&label=nuc%20mutations:T14257C

List of sequences:
EPI_ISL_14294806
EPI_ISL_14333750
EPI_ISL_14333754
EPI_ISL_14542445
EPI_ISL_14608324
EPI_ISL_14632425
England/PHEC-YYGIB6T/2022|2022-08-12
Originally posted by @FedeGueli in #931 (comment)
I decided to open an issue after just 7 sequences for three reason:
1-Convergent evolution at S:460 with BA.2.75
2-Already Sampled in 4 continents suggesting high or growing prevalence somewhere likely in Africa
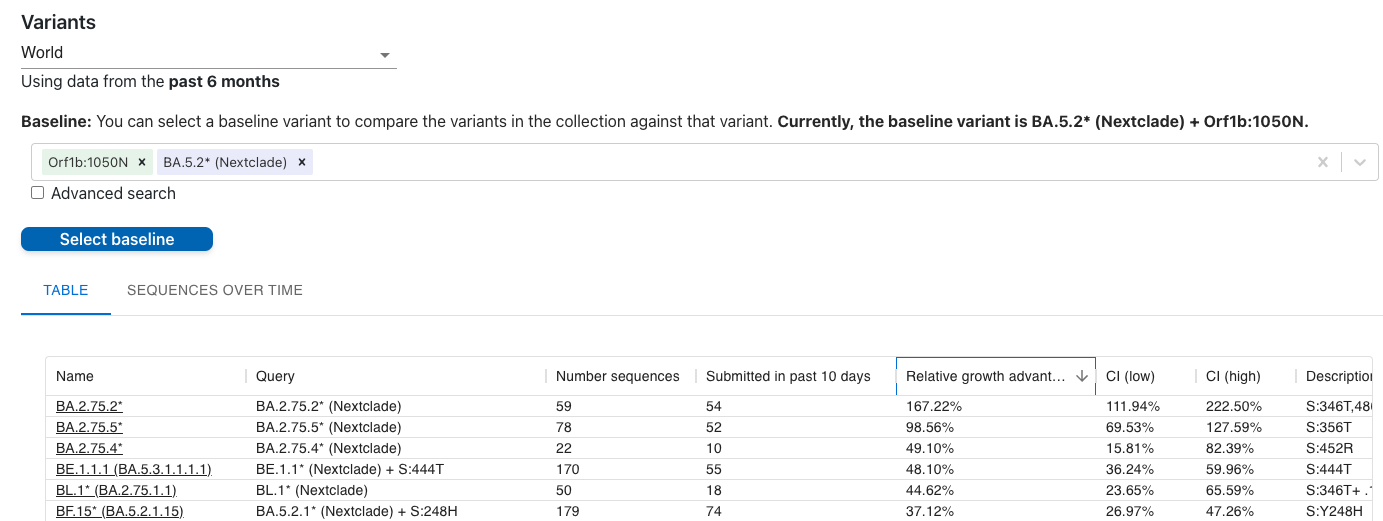
3-BE.1.1.1 is the fastest BA.5 sublineage according to our collection on covspectrum :
https://cov-spectrum.org/collections/24
:
The text was updated successfully, but these errors were encountered: