Abstract
Vaccines against SARS-CoV-2 have been shown to be safe and effective but their protective efficacy against infection in the brain is yet unclear. Here, in the susceptible transgenic K18-hACE2 mouse model of severe coronavirus disease 2019 (COVID-19), we report a spatiotemporal description of SARS-CoV-2 infection and replication through the brain. SARS-CoV-2 brain replication occurs primarily in neurons, leading to neuronal loss, signs of glial activation and vascular damage in mice infected with SARS-CoV-2. One or two doses of a modified vaccinia virus Ankara (MVA) vector expressing the SARS-CoV-2 spike (S) protein (MVA-CoV2-S) conferred full protection against SARS-CoV-2 cerebral infection, preventing virus replication in all areas of the brain and its associated damage. This protection was maintained even after SARS-CoV-2 reinfection. These findings further support the use of MVA-CoV2-S as a promising vaccine candidate against SARS-CoV-2/COVID-19.
Similar content being viewed by others
Main
Coronavirus disease 2019 (COVID-19) is caused by the infection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)1. Although COVID-19 is primarily a respiratory disease, many patients manifest neurological symptoms, including anosmia and ageusia, nonspecific symptoms such as headache or dizziness, or severe conditions such as cognitive impairment, epilepsy, ataxia or encephalopathy2. These symptoms have been attributed to the secondary effects of the systemic SARS-CoV-2 infection (that is, hypoxemia produced by severe pneumonia, cytokine release syndrome induced by hyperactivation of the immune response, thrombotic complications or electrolyte dysregulation by acute renal injury) or encephalitis produced by the direct viral infection of the central nervous system (CNS)3,4,5,6. However, direct CNS infection is supported by the neurotropism exhibited by other coronaviruses7,8 and by the detection of SARS-CoV-2 in cerebrospinal fluid from patients with COVID-19 and in a significant proportion of brain autopsies from patients who died from COVID-19 (refs. 3, 9, 10). Furthermore, SARS-CoV-2 has also been detected in the brain of different experimental animal models, including transgenic11,12 and knock-in mice13 expressing human angiotensin-converting enzyme 2 (hACE2) and natural hosts of SARS-CoV-2 such as hamsters6, ferrets14 and nonhuman primates15. Three main routes have been proposed by which SARS-CoV-2 may enter the CNS: (1) the so-called olfactory route, where the virus could reach the olfactory bulb directly through the lamina cribosa or by infection of olfactory sensory neurons6,16; (2) the hematological route, in which the virus enters the brain by crossing the blood–brain barrier (BBB) and/or blood–cerebrospinal fluid brain barrier; and (3) retrograde transport through peripheral nerves innervating the respiratory tract (that is, trigeminal, facial, glossopharyngeal and vagus nerves)17. Regardless of the pathogenic mechanism (viral neuroinvasion or secondary to the systemic infection) several studies have demonstrated important neuropathological alterations in patients with severe COVID-19, such as neurovascular pathology, glial activation and neuronal damage10,18,19,20. Additionally, biomarkers of cerebral injury have also been found to be elevated in patients with mild or moderate COVID-19 (ref. 21). Furthermore, neurological manifestations are common in patients recovered from the acute phase of COVID-19, suggesting the possibility of chronic brain impairment associated with the post-acute COVID-19 syndrome22,23.
Many vaccine candidates against COVID-19 have been developed and clinically tested in phase I, II and III trials. Vaccines approved by the main regulatory agencies are primarily based on the SARS-CoV-2 spike (S) protein and have been generated by various technologies including messenger RNA (Pfizer-BioNTech and Moderna)24,25, adenoviral vectors (AstraZeneca, Janssen and Sputnik)26,27,28 or inactivated virus (Sinopharm and Sinovac)29. These vaccines are currently being used for mass vaccination; however, it is still unknown whether they prevent viral spread to other regions of the body such as the CNS and confer protection against the brain damage induced by the SARS-CoV-2 infection. We have previously described the advantages of a poxvirus modified vaccinia virus Ankara (MVA) vector expressing a human codon-optimized, full-length SARS-CoV-2 S protein (termed MVA-CoV2-S) as a promising COVID-19 vaccine candidate. The MVA-CoV2-S vaccine candidate induces in mice robust and long-term memory S-specific humoral and T cell immune responses, and fully prevented morbidity, mortality, viral replication, pathology and cytokine storm in the lungs of K18-hACE2 transgenic mice infected with SARS-CoV-2 (refs. 30,31,32). Moreover, we have recently described that MVA-CoV2-S vaccination also induces a robust SARS-CoV-2-specific humoral and cellular immunogenicity and full efficacy against SARS-CoV-2 infection in other animal models, such as hamsters33 and rhesus macaques34.
Here, we examine the efficacy of MVA-CoV2-S vaccination to prevent SARS-CoV-2 cerebral infection and associated damage in K18-hACE2 mice, a well-established mouse model of severe COVID-19 disease11,12,35. To this end, we provide a detailed spatiotemporal description of the SARS-CoV-2 viral spread among the main regions of the brain. SARS-CoV-2 infection and replication appear mainly restricted to neurons, producing significant neuronal cell death. Indeed, as described previously19, infected mice also exhibit pathological alterations in brain blood vessels. Administration of one or two doses of the MVA-CoV2-S vaccine candidate confers full protection against SARS-CoV-2 neuroinvasion, preventing cerebral viral replication and the associated brain damage, even after reinfection. This supports that MVA-CoV2-S is a promising vaccine candidate against SARS-CoV-2/COVID-19.
Results
Characterization of SARS-CoV-2 brain infection in K18-hACE2 mice
SARS-CoV-2 is transmitted by exposure to the nasal or oral cavity and primarily replicates along the respiratory tract producing, in severe cases, pulmonary disease. Furthermore, SARS-CoV-2 can disseminate into the circulation and in this way can infect other organs such as kidney, heart, brain or the gastrointestinal tract2. Although SARS-CoV-2 CNS infection has been well described in susceptible transgenic K18-hACE2 mice3,4,12,36,37,38, little information about viral spreading to specific cerebral areas has been reported. Thus, to study in detail the spatiotemporal SARS-CoV-2 viral distribution and replication in the brain, K18-hACE2 mice (n = 26, 11 females and 15 males) were inoculated intranasally with SARS-CoV-2 (MAD6 isolate, 1 × 105 plaque-forming units (PFU) per mouse)30,31 and their brains were examined by immunohistochemistry against SARS-CoV-2 nucleocapsid (N) protein at 2 (n = 8), 4 (n = 8) and 6 (n = 10) days postinfection (dpi) (Fig. 1a–c and Extended Data Fig. 1a). At 6 dpi all mice infected with SARS-CoV-2 lost more than 25% of body weight due to severe pulmonary disease (with decreased gas exchange, plasma electrolyte dysregulation and systemic cytokine/chemokine storm) and to the significant brain infection causing encephalitis4,11,12,30,31,32, and were euthanized. Figure 1a shows brain coronal sections from representative control (uninfected; n = 9, 4 females and 5 males) and SARS-CoV-2-infected mice (6 dpi) revealing that the SARS-CoV-2 N staining was clear and specific, with many infected cells throughout different regions of the brain. The precise analysis of the brain viral distribution at different time points, which did not reveal any sex-based differences, is shown in Fig. 1b,c and Extended Data Figs. 1a and 2a,b. At 2 dpi, no evidence of SARS-CoV-2 infection was found in any of the brain areas studied in the 8 mice analyzed. At 4 dpi, variable levels of viral infection were observed in the different cerebral regions examined in the 8 mice analyzed. Specifically, the basal forebrain, amygdala and hypothalamus showed the highest levels of SARS-CoV-2 N staining at this time point, with many groups of SARS-CoV-2-infected cells in most of the brains analyzed. In other regions, such as the olfactory bulb, cortex, or mesencephalon, an intermediate level of infection was detected, with only some dispersed infected cells in most of the brains studied. In some mice, regions like the striatum, different areas of the hippocampus, thalamus, pons and cerebellum showed a few SARS-CoV-2+ cells, indicating the lower level of infection at 4 dpi. Finally, at the latest time point studied, 6 dpi, all brains analyzed (n = 10) revealed high levels of SARS-CoV-2 N staining, but showed a nonhomogeneous distribution of viral infection among the main areas of the brain. In the olfactory bulbs, cortex, basal forebrain, amygdala, thalamus, hypothalamus and mesencephalon, a severe SARS-CoV-2 infection was detected. Other regions, such as hippocampal CA1 and dentate gyrus and pons, showed moderate infection; whereas in the striatum, the CA2/CA3 area of the hippocampus and cerebellum, only some disperse SARS-CoV-2+ cells were detected suggesting a mild viral infection.
a, Brain coronal sections of control (uninfected) and SARS-CoV-2-infected K18-hACE2 mice (6 dpi) after immunohistochemistry against the SARS-CoV-2 N protein. b, Qualitative analysis of SARS-CoV-2 level of infection in different cerebral regions of infected K18-hACE2 mice at 2, 4 and 6 dpi. Between brackets, the number of mice showing SARS-CoV-2+ cells among the total number of mice studied is indicated for each brain region analyzed. c, High-magnification images, after SARS-CoV-2 N immunohistochemistry, illustrating the time course of SARS-CoV-2 infection in the cortex, hypothalamus and mesencephalon of control and infected K18-hACE2 mice. SARS-CoV-2 N immunostaining was performed in six independent experiments obtaining similar results. d, Quantitative analysis of SARS-CoV-2 RNA, detected by RT–qPCR targeting the viral E gene, in the olfactory bulb, cortex, hypothalamus and brain stem. Data are presented as the mean ± s.e.m. SARS-CoV-2-infected mice: 2 dpi, n = 8 (3 females and 5 males); 4 dpi, n = 8 (3 females and 5 males); and 6 dpi, n = 10 (5 females and 5 males) mice. Kruskal–Wallis test, post hoc Dunn’s test. *P < 0.05; ***P < 0.001; ****P < 0.0001 with regard to the RT–qPCR values obtained, in each respective brain region, of uninfected controls.
SARS-CoV-2 infection in K18-hACE2 mice produces anosmia35 and different authors have proposed olfactory bulb infection as the principal route of neuroinvasion4,6,15. To advance in knowledge on the route of viral entry in the CNS, we studied the neurotropism, that is the ability of the virus to infect and replicate in CNS regions17, by highly sensitive quantitative reverse transcription PCR (RT–qPCR) of the SARS-CoV-2 subgenomic E gene at 2, 4 and 6 dpi. RT–qPCR analyses were done in (1) the olfactory bulb, the target of the olfactory route6,15,16, (2) the cortex and hypothalamus, as two representative potential targets of the hematological route (the cortex with a highly restrictive BBB and the hypothalamus with areas of less restrictive BBB39), and (3) the brain stem, as the target of sensory fibers innervating the respiratory tract. At 2 dpi, only minimal levels of SARS-CoV-2 subgenomic RNA were found in the different brain regions analyzed; no statistically significant differences were seen in any of these brain areas compared to each other or with regard to uninfected controls (Fig. 1d and Extended Data Fig. 1b). At 4 dpi, similar levels of SARS-CoV-2 subgenomic RNA were found between samples from the four brain areas analyzed, showing statistically significant differences with regard to uninfected controls in the olfactory bulb, cortex and hypothalamus, as well as a small difference (P = 0.055) in the brain stem (Fig. 1d). At 6 dpi, increased levels of SARS-CoV-2 subgenomic RNA were found in all samples studied, being the cortex and hypothalamus the structures with the highest levels of viral RNA (Fig. 1d). Given that we have not observed earlier or higher viral replication in the olfactory bulb or brain stem in comparison with other brain areas, it can be suggested that, as expected, neither the olfactory route nor the retrograde transport from respiratory innervation are the main port of cerebral viral entry. It seems that the hematological route is the predominant route of SARS-CoV-2 infection in the brain of K18-hACE2 mice.
An important observation, revealed by the histological analyses of brains from SARS-CoV-2-infected mice, is that most of the infected cells show a neuronal morphology (Fig. 1c and Extended Data Figs. 1a and 2a), suggesting that viral replication occurs primarily in neurons. This was confirmed by high-resolution confocal microscopy analysis combining SARS-CoV-2 N protein immunofluorescence with neuronal (neuronal-specific nuclear antigen A60 (NeuN)), astroglial (glial fibrillary acid protein (GFAP)), microglial (ionized calcium-binding adapter molecule 1 (IBA1)) and vascular endothelial (isolectin B4 (IB4)) markers in SARS-CoV-2-infected brains at 6 dpi (Fig. 2 and Extended Data Fig. 4). As indicated in Fig. 2a, all cells showing high SARS-CoV-2 staining (green) were also positive for the neuronal marker NeuN (red). In addition, a confocal orthogonal projection confirmed that both SARS-CoV-2+ and NeuN+ signals colocalized in the same confocal plane (Z-depth resolution of confocal plane = 0.7 μm; Fig. 2b), indicating that the SARS-CoV-2 N protein and the NeuN protein are within the same neuronal body. SARS-CoV-2 infection and replication appear to take place in a broad variety of neuronal subtypes; we found high levels of SARS-CoV-2 N protein staining in cortical glutamatergic-Ca2+/calmodulin-dependent protein kinase-II (CaMKII)+ and GABAergic-parvalbumin+ neurons, striatal DARPP32+ medium spiny neurons and parvalbumin+ interneurons, cholinergic-choline acetyltransferase (ChAT)+ neurons of the basal forebrain and mesencephalic and hypothalamic dopaminergic-TH+ neurons (Fig. 2c–f and Extended Data Fig. 3). SARS-CoV-2+ infection of nonneuronal cells was evaluated by confocal microscopy analysis combining SARS-CoV-2 N protein staining with microglial (IBA1), astroglial (GFAP) or vascular (IB4) markers (Extended Data Fig. 4). We did not observe colocalization of SARS-CoV-2+ and GFAP+ staining, indicating the absence of viral particles in astrocytes (Extended Data Fig. 4a–c). In contrast, in some vascular cells (IB4+), discrete SARS-CoV-2+ staining was detected, suggesting SARS-CoV-2 infection in brain blood vessels, as reported previously19 (Extended Data Fig. 4d). Furthermore, our analysis also showed many microglial cells with processes contacting or engulfing SARS-CoV-2-infected neurons or damaged vessels (Extended Data Fig. 4e–g). In some cases, we could even detect SARS-CoV-2+ staining inside IBA1+ cells (Extended Data Fig. 4h), suggesting that viral particles from infected neurons or damaged vascular cells may have been phagocyted by microglial cells.
a, High-resolution Z-projection confocal images of the cortex of SARS-CoV-2-infected K18-hACE2 mice, after SARS-CoV-2 N protein (green) and NeuN (red) immunofluorescence detection, showing specific strong viral load in neuronal cells. b, Orthogonal projection of the cortical neuron indicated by an arrow in a, demonstrating the colocalization of the cytoplasmic SARS-CoV-2 and nuclear NeuN signals in the same neuronal cell. The confocal image corresponds to a single confocal plane (z = 10) of 661 nm Z-depth optic resolution. c–f, Microscopy confocal images showing the colocalization of SARS-CoV-2 N protein (green) with the neuronal subtype-specific markers (red): CaMKII (c), parvalbumin (d), ChAT (e) and tyrosine hydroxylase (f), showing the viral infection of cortical glutamatergic-CaMKII+ and GABAergic-parvalbumin+ neurons, cholinergic-ChAT+ neurons of the basal forebrain and mesencephalic dopaminergic-tyrosine hydroxylase+ neurons. The insets are depicted at higher magnification on the right. The arrows label some examples of SARS-CoV-2-infected neurons. Nuclei were counterstained with DAPI (blue). Immunofluorescence staining was performed in 3 independent experiments obtaining similar results, analyzing SARS-CoV-2-infected mice at 6 dpi, n = 6 (3 females and 3 males). In e, AC, anterior commissure.
Taking these results together, our study indicates that SARS-CoV-2 brain replication in K18-hACE2 mice occurs primarily in neurons, beginning between 2 and 4 d after inoculation with SARS-CoV-2, with the highest levels of infection seen in ventral areas of the brain, such as the hypothalamus, amygdala, and basal forebrain. In a later phase, between 4 and 6 dpi, viral replication spreads to most cerebral regions, producing a severe SARS-CoV-2 infection. Interestingly, even at 6 dpi, some specific cerebral areas, such as the cerebellum, striatum and CA2/CA3 region of the hippocampus, remain with mild levels of SARS-CoV-2 infection, presenting only some dispersed SARS-CoV-2-infected neurons.
Neuropathological alterations associated with SARS-CoV-2 brain infection
Next, we studied whether a strong SARS-CoV-2 infection induces neuronal death by analyzing the neuronal density in the hypothalamus and cortex, two areas with high viral replication, and in the hippocampus, which present mild-to-moderate viral infection. The stereological quantification of hypothalamic NeuN+ (Fig. 3a,b) and cortical Nissl+ (Fig. 3c,d) neurons demonstrated a significant decrease of neuronal density in SARS-CoV-2-infected mice at 4 and 6 dpi, compared to uninfected control mice. Moreover, a significant loss of NeuN+ neurons was also detected in the hippocampal dentate gyrus at 6 dpi, whereas no differences with regard to uninfected controls were detected in the CA1 or CA2/CA3 regions (Extended Data Fig. 2c). Since SARS-CoV-2 infection can induce neuronal apoptosis in human brain organoids5, we studied by immunodetection the number of cells expressing cleaved caspase-3 (c-casp3) in brains from control (uninfected) and SARS-CoV-2-infected mice at 4 and 6 dpi. As expected, the brains of control mice showed only few c-casp3+ cells in the hippocampus (Fig. 3e,f), possibly reflecting physiological apoptosis associated with the neurogenic niche of the dentate gyrus40; practically no c-casp3+ cells were detected in the rest of the brain (Fig. 3g,h and Extended Data Fig. 5). In contrast, brains of SARS-CoV-2-infected mice presented a substantial number of c-casp3+ cells distributed across most of the brain areas analyzed, being particularly evident at 6 dpi (Fig. 3e–h and Extended Data Fig. 5), when the brain viral infection is maximal. The distribution of c-casp3+ cells suggests that a significant proportion of apoptotic cells correspond to neurons. Quantitative analyses of apoptotic cell numbers were performed in the hippocampus (Fig. 3f) and hypothalamus (Fig. 3h). In both regions, we found a statistically significant increase in the number of c-casp3+ cells in SARS-CoV-2-infected mice.
a–d, Light-transmitted images of hypothalamic NeuN+ (a) and cortical Nissl+ (c) neurons of control and SARS-CoV-2-infected mice and stereological quantification of neuronal density in these areas (b,d). e–h, Hippocampal (e) and hypothalamic (g) images from control and SARS-CoV-2-infected mice after immunohistochemistry against apoptotic c-casp3. Images in the right column depict, at higher magnification, the insets of the central column. f, Quantification of the density of c-casp3+ cells in the hippocampus. h, Quantification of the density of c-casp3+ cells in the hypothalamus. In b,d,f,h, data are presented as the mean ± s.e.m. Control mice, n = 9 (4 females and 5 males); SARS-CoV-2-infected mice, 4 dpi, n = 8 (3 females and 5 males); and 6 dpi, n = 10 (5 females and 5 males). ANOVA with post hoc Dunnett’s test. b,d,f: b, 4 dpi: P = 0.003, 6 dpi: P < 0.0001; d, 4 dpi: P = 0.0004, 6 dpi: P < 0.0001; f, 6 dpi: P = 0.0003). h, Kruskal–Wallis test with post hoc Dunn’s test. **P < 0.01; ***P < 0.001; ****P < 0.0001, with regard to controls (uninfected mice).
Morphological changes in microglia and structural alterations in cerebral blood vessels have been described in patients with COVID-19 (refs. 19, 20). Thus, to study the glial reaction in brains from K18-hACE2 mice infected with SARS-CoV-2, we analyzed the presence of astrogliosis and reactive microglia using specific immunostaining of astroglial (GFAP) and microglial (IBA1) markers. The qualitative analysis of GFAP+ fluorescence signal did not reveal clear signs of astrogliosis, either by GFAP overexpression or by significant morphological changes in GFAP+ astrocytes, in any of the cerebral regions studied (cortex, hippocampus and hypothalamus; Extended Data Fig. 6, Fig. 4 and below). However, IBA1 immunostaining in the same brain regions showed some morphological changes suggesting microglial activation (Extended Data Fig. 6). Microglial activation is characterized by marked morphological changes that include enlargement of the cell body and an important retraction of projections, resulting in a more ameboid shape. The quantitative analysis of the microglial morphology (using the Imaris microscopy image analysis software) in the cortex and hypothalamus revealed changes in SARS-CoV-2-infected mice at 6 dpi, with a significant reduction in the microglial total and filament area, filament length and number of filaments branching points (Fig. 4a–d). We also analyzed the density of microglia, astrocytes and oligodendrocytes at 6 dpi, when viral infection is maximal. An increased number of IBA1+ microglial cells, with regard to uninfected controls, was found in the cortex and hypothalamus (Fig. 4e); the density of GFAP+ astrocytes was also increased in the hypothalamus, with a nonsignificant trend to higher density in the cortex (Fig. 4f), whereas the density of oligodendrocytes in the corpus callosum was not altered (Fig. 4g). Taken together, these data indicate that at 6 dpi (the stage at which mice were euthanized due to a marked loss of body weight) the microglial and astrocytic responses induced by SARS-CoV-2 infection were still in an early stage, although clear signs of microglial activation were already observed.
a–d, Representative images of Imaris 3D reconstruction of cortical (a) and hypothalamic (c) microglial cells from uninfected controls and SARS-CoV-2-infected mice (6 dpi) and quantitative morphological analysis of microglial cells in the cortex (b) and hypothalamus (d) showing total area, filament area and length and number of filament branching points. e–g, Quantification of the density of IBA1+ microglial cells (e) and GFAP+ astrocytes (f) in the cortex and hypothalamus and oligodendrocytes in the corpus callosum (g). h, High-magnification confocal images from control and SARS-CoV-2-infected K18-hACE2 mice stained with the vascular marker IB4 (red), showing the alterations observed in the brain vessels of SARS-CoV-2-infected animals. i, Analysis of the presence of abnormal blood vessels in the hypothalamus of controls and SARS-CoV-2-infected mice at 2, 4 and 6 dpi. In b,d,e,f,g,i, data are presented as the mean ± s.e.m. b,d, Representation of the individual microglial cell values from: control mice, n = 9 (4 females and 5 males); SARS-CoV-2-infected mice: 2 dpi, n = 8 (3 females and 5 males); 4 dpi, n = 8 (3 females and 5 males); and 6 dpi, n = 10 (5 females and 5 males). e–g, Control mice, n = 9 (4 females and 5 males (e,f)) and n = 8 (3 males and 5 males (g)); SARS-CoV-2-infected mice, 6 dpi, n = 9 (4 females and 5 males). i, Control mice, n = 9 (4 females and 5 males); SARS-CoV-2-infected mice: 2 dpi, n = 8 (3 females and 5 males); 4 dpi, n = 8 (3 females and 5 males); and 6 dpi, n = 10 (5 females and 5 males). Kruskal–Wallis test with post hoc Dunn’s test (b,d,i) and unpaired, two-sided t-test (e,f,g). In e,f: e, cortex: P = 0.001, hypothalamus: P < 0.0001; f, hypothalamus: P < 0.0001). **P < 0.01, ***P < 0.001, ****P < 0.0001 with regard to controls (uninfected mice).
To study the presence of vascular pathology induced by SARS-CoV-2 infection, brain blood vessels were labeled with IB4, a marker of the luminal and abluminal side of endothelial cells, and vessel abnormality was evaluated as described previously41. No significant alterations in the cerebral blood vessels of infected mice were found at 2 and 4 dpi. However, at 6 dpi, when brain viral infection was maximal, histological evidences of abnormal blood vessels started to appear in ventral brain areas (basal forebrain, amygdala and hypothalamus; Fig. 4h,i and Extended Data Fig. 6), which, in some cases, extended to other heavily infected regions. The endothelial cells became round by losing their elongated morphology, IBA1+ innate immune cells covered the vessels and presented phagocytic pouches including IB4+ material (Extended Data Fig. 4g), suggesting inflammatory activation of vessel permeability and remodeling, similar to that observed in experimental models of multiple sclerosis41. Interestingly, as in other inflammatory models, vascular damage was restricted to arterioles, the vascular region more susceptible to immune cells crossing under inflammation. These results agree with the vascular brain pathology described in patients with COVID-19, hamsters and K18-hACE2 mice infected with SARS-CoV-2 (ref. 19). They also indicate that SARS-CoV-2 infection in the K18-hACE2 mouse model of severe COVID-19 produces important neuropathological alterations, including neuronal loss, incipient signs of gliosis and vascular damage.
MVA-CoV2-S vaccination fully prevents SARS-CoV-2 brain infection and damage
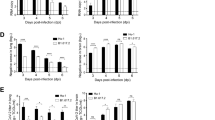
Once the temporal and regional spread of SARS-CoV-2 and associated neuropathology were characterized in the brain of K18-hACE2 mice, we next tested whether the vaccine candidate MVA-CoV2-S (also termed MVA-S), expressing the SARS-CoV-2 S protein30, could protect against SARS-CoV-2 brain infection and associated damage. Thus, K18-hACE2 mice were immunized by intramuscular route with 1 or 2 doses of MVA-S (1 × 107 PFU per mouse) at days 0 and 28; subsequently, on day 63, they were challenged with a lethal intranasal dose of SARS-CoV-2 (MAD6 isolate; 1 × 105 PFU per mouse), as reported previously30,31,32. SARS-CoV-2-challenged mice primed and boosted with MVA-wild type (WT) (WT empty MVA vector) were used as positive control of infection (Fig. 5a, upper panels). Then, at 4 dpi (day 67), mice were euthanized for brain extraction and processing (MVA-WT, n = 9; MVA-S 1 dose, n = 9; MVA-S 2 doses, n = 3). Moreover, in a second independent experimental approach, we evaluated whether mice vaccinated with one or two doses of MVA-S, which survived SARS-CoV-2 infection30, were protected against viral neuroinvasion after a SARS-CoV-2 reinfection performed 46 d after the first SARS-CoV-2 challenge. In this experiment, SARS-CoV-2-challenged unvaccinated and MVA-WT-inoculated mice were used as positive controls of infection (Fig. 5a, lower panels). Thereafter, mice (MVA-WT, n = 5; SARS-CoV-2, n = 4; MVA-S 1 dose, n = 5; MVA-S 2 doses, n = 4) were euthanized for brain extraction and processing 6 days after the second viral infection (6 dpi) or 6 d after the first infection in the unvaccinated and MVA-WT groups. In both experimental approaches, the presence of cerebral SARS-CoV-2 infection was analyzed by immunohistochemistry against the SARS-CoV-2 N protein in different brain regions, as described above. Interestingly, all MVA-S vaccinated mice, either with one or two doses, showed total protection against cerebral SARS-CoV-2 infection after a single SARS-CoV-2 infection (Fig. 5b,c) or after a reinfection (Fig. 5c and Extended Data Fig. 7), without any SARS-CoV-2+ infected cells being detected in any of the brain regions analyzed. The absence of SARS-CoV-2+ immunostaining observed in MVA-S-vaccinated mice contrasts with the high number of SARS-CoV-2+ infected cells found in challenged MVA-WT-inoculated mice (Fig. 5b,c and Extended Data Fig. 7) or in challenged unvaccinated mice (Fig. 5c and Extended Data Fig. 7). Furthermore, to discard the possibility that the absence of SARS-CoV-2+ labeling in MVA-S-vaccinated mice was due to a low viral load, which could be below the immunohistochemistry detection limit, we performed highly sensitive RT–qPCR of the SARS-CoV-2 E gene in the cortex and hypothalamus, two brain regions that present high viral replication. According to the histological analysis, high levels of SARS-CoV-2 subgenomic mRNA were found in the cortex and hypothalamus of SARS-CoV-2-challenged MVA-WT or unvaccinated mice (Fig. 5d,e). Importantly, SARS-CoV-2 subgenomic mRNA was not detected in any of the MVA-S-vaccinated mice, regardless of the vaccination regimen (one or two doses) or whether they were subjected to a single SARS-CoV-2 infection (Fig. 5d) or reinfection (Fig. 5e). As described previously, the protection against SARS-CoV-2 infection observed in MVA-S vaccinated mice correlated with the high titers of SARS-CoV-2 neutralizing antibodies induced after immunization (about 4 × 102 and 2 × 103 NT50 titers in mice vaccinated with 1 and 2 doses, respectively), with neutralizing antibody titers above 3 × 102 inducing protection from lung infection31. In MVA-S-vaccinated mice these neutralizing antibody titers were maintained at late times after the first SARS-CoV-2 viral infection31,32; in the case of mice vaccinated with two doses of MVA-S similar high titers of SARS-CoV-2 neutralizing antibodies were induced before and after the first and second viral infections, indicating the lack of breakthrough infection31,32. Together, these results clearly demonstrate that MVA-S vaccination confers complete and sustained protection against SARS-CoV-2 cerebral infection.
a, Schematic diagram showing the schedule used to analyze the protection induced by MVA-CoV2-S vaccination against SARS-CoV-2 brain infection in K18-hACE2 mice. Note that MVA-CoV2-S was tested for a single SARS-CoV-2 infection (top, brain extraction at 4 dpi) or for a second reinfection (bottom, brain extraction at 6 dpi). b, High-magnification images from the cortex, hypothalamus and mesencephalon after SARS-CoV-2 N immunohistochemistry, illustrating the efficacy of the MVA-S vaccine candidate, in a regimen of one or two doses, against SARS-CoV-2 cerebral infection. SARS-CoV-2 N immunostaining was performed in five independent experiments obtaining similar results. c, Qualitative analysis of the level of SARS-CoV-2 infection in the different cerebral regions of the experimental groups showed in a. The number of mice showing SARS-CoV-2+ cells among the total number of mice studied is indicated for each brain region analyzed and shown in brackets. d–e, Quantitative analysis of SARS-CoV-2 RNA detected by RT–qPCR targeting the viral E gene, in the cortex and hypothalamus, of the experimental groups described previously for the SARS-CoV-2 infection (d) or reinfection (e) experiments. Data are presented as the mean ± s.e.m. SARS-CoV-2 infection experiment (4 dpi): MVA-WT, n = 9 (6 males and 3 females); MVA-S 1 dose, n = 9 (6 males and 3 females); MVA-S 2 doses, n = 3 females. SARS-CoV-2 reinfection experiment (6 dpi): MVA-WT, n = 5 females; SARS-CoV-2, n = 4 females; MVA-S 1 dose, n = 5 females; MVA-S 2 doses, n = 4 females. Kruskal–Wallis test with post hoc Dunn’s test (d,e). *P < 0.05; **P < 0.01; ***P < 0.001, with regard to MVA-WT- or SARS-CoV-2-infected mice.
We also evaluated the efficacy of MVA-S vaccination to protect against brain damage induced by severe SARS-CoV-2 infection. As expected by the absence of viral infection in MVA-S-vaccinated mice, the stereological quantification of the density of hypothalamic NeuN+ (Fig. 6a) and cortical Nissl+ (Fig. 6b) neurons clearly demonstrated that MVA-S vaccination, either with one or two doses, protects against neuronal death induced by the encephalitis caused by SARS-CoV-2 infection. Furthermore, analysis of apoptotic cells revealed the absence of c-casp3+ cells in all brains of MVA-S-vaccinated mice, with the exception of physiological hippocampal apoptosis also detected in uninfected mice (data not shown). Quantification of the number of c-casp3+ cells in the hypothalamus confirmed that MVA-S vaccination confers a complete protection against CNS cellular apoptosis induced by SARS-CoV-2 infection (Fig. 6c). The quantitative morphological analysis of microglial IBA1+ cells revealed morphological changes compatible with microglial activation in MVA-S-vaccinated mice (Fig. 6d,e and Extended Data Fig. 8), despite the total absence of cerebral infection observed in these animals (Fig. 5c,e and Extended Data Fig. 7). Moreover, microglial morphological changes in vaccinated mice were more pronounced in the hypothalamus (Fig. 6d and Extended Data Fig. 8), with a permeable BBB, and after two doses of the MVA-S vaccine, which induce a stronger immune response31,32, suggesting that the systemic immune response induced by MVA-S vaccination also activated brain-resident immune cells, reinforcing the idea of active communication between the peripheral and central compartments of the immune system42. Furthermore, the analysis of brain blood vessels after IB4 staining also showed protection in MVA-S-vaccinated mice against the appearance of abnormal brain blood vessels after SARS-CoV-2 infection (Fig. 6f and Extended Data Fig. 8).
a,b, Stereological estimation of hypothalamic NeuN+ (a) and cortical Nissl+ neurons (b) from the control (uninfected) and experimental groups of the reinfection experiment (6 dpi): SARS-CoV-2 and MVA-S, 1 and 2 doses. c, Quantification of apoptotic c-casp3+ cells in the hypothalamus of SARS-CoV-2 and MVA-S (one or two doses) mice used in the reinfection experiment. d,e, Quantitative morphological analysis of microglial cells, using the Imaris software, in the cortex (b) and hypothalamus (d) showing total area, filament area and length and number of filament branching points from the experimental groups described. f, Quantification of abnormal blood vessel density in the hypothalamus. Data are presented as the mean ± s.e.m. Controls, n = 4 females; SARS-CoV-2, n = 4 females; MVA-S: 1 dose, n = 5 females; MVA-S 2 doses, n = 4 females (a–e) and n = 3 females (f). d,e, Representation of the individual microglial cell values from the experimental mice exposed as described above. ANOVA with post hoc Dunnett’s test (a, control versus SARS-CoV-2: P = 0.0003; SARS-CoV-2 versus MVA-S 1 dose: P = 0.0003; SARS-CoV-2 versus MVA-S 2 doses: P = 0.0004) or Fisher’s LSD (b, control versus SARS-CoV-2: P = 0.0001; SARS-CoV-2 versus MVA-S 1 dose: P = 0.0002; SARS-CoV-2 versus MVA-S 2 doses: P = 0.0003) and Kruskal–Wallis test with post hoc Dunn´s test (c,d,e,f). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, with regard to SARS-CoV-2-infected mice.
Taken together, these data demonstrate that MVA-S vaccination confers a complete protection against SARS-CoV-2 brain infection and the associated neuropathological damage (neuronal loss and vascular damage), even after a second viral infection. Interestingly, cerebral protection induced by the MVA-S vaccine candidate is achieved similarly with one or two doses.
Discussion
After respiratory symptoms, neuropsychiatric manifestations are the second most common symptoms in patients with COVID-19 (refs. 2, 26). Despite the clinical relevance of the brain damage caused by COVID-19, it is still unknown whether the different COVID-19 vaccine candidates can prevent SARS-CoV-2 neuroinvasion or associated damage. In this study, we show that a vaccine candidate against COVID-19 based on the poxvirus MVA vector expressing the SARS-CoV-2 S protein (MVA-CoV2-S) confers complete protection against SARS-CoV-2 brain infection. To test the efficacy of the MVA-S vaccine candidate, we used the K18-hACE2 mouse model of severe COVID-19 (refs. 11, 12). This transgenic mouse model has increased hACE2 cerebral expression43, presenting significant brain permissiveness to SARS-CoV-2 replication. Our histological analysis revealed that ventral areas of the brain (basal forebrain, hypothalamus and amygdala) are the first cerebral regions infected by SARS-CoV-2, with virus replication being detected at 4 dpi. On the other hand, the olfactory bulbs, which have been proposed as one of the main ports of the viral CNS entry6,15,16, presented mild SARS-CoV-2 infection at 4 dpi and only showed severe viral infection after 6 dpi, when SARS-CoV-2 replication had spread to most of the brain regions. The molecular study of viral neurotropism failed to show earlier or higher levels of viral replication in the olfactory bulb. These data are consistent with recent studies that failed to detect significant levels of viral replication in the olfactory bulbs of patients who died a few days after viral infection44. In addition, the fact that the hypothalamus, where there are highly fenestrated BBB capillaries39, is one of the brain regions with the highest and earliest viral replication levels, suggesting that the hematogenous is the main route of entry of SARS-CoV-2 into the CNS45. Another relevant finding of our analysis of SARS-CoV-2 infection in K18-hACE2 mice is that brain viral replication occurs primarily in neurons, inducing significant neuronal cell death. These findings are consistent with the detection of SARS-CoV-2 in cortical neurons from deceased patients with COVID-19 and with the induction of neuronal apoptosis in infected human brain organoids5.
Our study clearly demonstrates that MVA-CoV2-S vaccination confers sterilizing immunity against brain viral replication and damage. In previous studies, we reported that the MVA-CoV2-S vaccine candidate induced in mice robust SARS-CoV-2-specific humoral and cellular immune responses, producing high titers of binding IgG antibodies against the S and receptor-binding domain proteins, high titers of neutralizing antibodies able to recognize different variants of concern and potent, broad and polyfunctional S-specific T cell immune responses30,31,32. Moreover, memory SARS-CoV-2-specific humoral and cellular immune responses were detected in mice even at six months after the last MVA-CoV2-S immunization31. We have also established that K18-hACE2 mice vaccinated with MVA-CoV2-S and challenged with SARS-CoV-2 are protected against mortality, body weight loss, viral lung replication and lung pathology and have reduced levels of pro-inflammatory cytokines, the two-dose treatment being more effective that one single dose30,31,32.
SARS-CoV-2 replication in K18-hACE2 mice is well described to occur primarily in the respiratory tract, during the first 2–4 dpi, and later on the cerebral tissue, between 3 and 7 dpi11,12. Probably, the exhaustive control exerted by MVA-CoV2-S vaccination on viral replication on the respiratory tract prevents neuroinvasion. The fact that immunization with a single dose of MVA-CoV2-S reduces but does not prevent virus infection in the lungs30,31,32, contrasts with the complete inhibition of brain viral infection in mice vaccinated with a single dose reported in this study and suggest that the block of viral brain infection could be due to the broad specificity of the immune responses triggered by MVA-CoV2-S vaccination. This inhibition is probably the result of the combined action of SARS-CoV-2-specific neutralizing antibodies and of CD4+ and CD8+ T cell responses triggered by vaccination, in turn preventing virus access to the brain. Thus, the relevance of vaccine protection from brain infection is an important requirement to block the spread of virus infection in tissues, long-term COVID-19 and mortality. Hence, vaccines that prevent SARS-CoV-2 infection in the brain of susceptible animals should be an indicator for vaccine development against variants of concern and of attenuated variants, like Omicron46.
To the best of our knowledge, only three articles have addressed the efficacy of COVID-19 vaccine candidates to protect against SARS-CoV-2 cerebral infection. In these works, the efficacy of adenoviral47, lentiviral48 or vesicular stomatitis virus49 S-based vaccines against SARS-CoV-2 brain infection were analyzed using K18-hACE2 transgenic mice, obtaining different outcomes. The adenoviral S-based vaccine candidate failed to control the SARS-CoV-2 brain replication, reducing the brain viral load only when it was combined with a nucleocapsid-based vaccine candidate, whereas the lentiviral or vesicular stomatitis virus S-based vaccine candidates were able to block the SARS-CoV-2 cerebral replication. Interestingly, our MVA-CoV2-S vaccine candidate not only completely abolished SARS-CoV-2 brain replication, even with one single dose, but also conferred sustained protection against a second viral infection, all vaccinated mice being completely resistant to a SARS-CoV-2 reinfection seven weeks after the first challenge. Interestingly, MVA-CoV2-S was able to induce memory SARS-CoV-2-specific humoral and CD4+ and CD8+ T cell immune responses even six months after the last dose31, strengthening the potent immunogenicity and durability of this vaccine candidate.
An important aspect of our data is that MVA-CoV2-S vaccination conferred complete protection against the cerebral damage induced by a severe SARS-CoV-2 infection, independently of the one- or two-dose vaccination regimes, with no evidence of cellular apoptosis, neuronal death or vascular alterations in any of the vaccinated mice. In a very stringent COVID-19 model as the K18-hACE2, where SARS-CoV-2 neurotropism increases, most of the neuropathological alterations induced during viral infection should be produced by direct viral neuroinvasion3,4. Therefore, the complete protection exerted by the MVA-CoV2-S vaccine candidate against cerebral SARS-CoV-2 infection and replication should be the main cause of the lack of neuropathological signs observed in the brains of vaccinated mice. Furthermore, the cytokine and chemokine storm produced by the systemic SARS-CoV-2 infection in many patients with COVID-19 has also been proposed to induce cerebral damage, producing neurological symptoms50. In this regard, we previously reported that MVA-CoV2-S vaccination prevented in K18-hACE2 mice the increase in pro-inflammatory cytokines induced by SARS-CoV-2 infection31,32, helping to reduce the potential cytokine-induced neurotoxicity in vaccinated K18-hACE2 mice.
In summary, this study shows that the MVA-CoV2-S vaccine candidate confers complete and sustained protection against SARS-CoV-2 brain infection, replication and the associated damage. These results, together with the previously described potent immunogenicity and full efficacy of MVA-CoV2-S in different animal models30,31,32,33,34, support the evaluation of this COVID-19 vaccine candidate in clinical trials.
Methods
Animals
Transgenic female and male K18-hACE2 mice, expressing the human ACE2 gene, were obtained from the Jackson Laboratory (034860-B6.Cg-Tg(K18-ACE2)2Prlmn/J, genetic background C57BL/6J × SJL/J)F2; research resource identifier (RRID): IMSR_JAX:034860). Experiments were carried out at the biosafety level 3 facilities of the Centro de Investigación en Sanidad Animal (CISA)-Instituto Nacional de Investigaciones Agrarias (INIA)-Consejo Superior de Investigaciones Científicas (CSIC) (Madrid, Spain). Mice were housed at 22 ± 1 °C under a 12-h light–dark cycle, with ad libitum access to food and water. Animal experimentation was approved by the Ethical Committee of Animal Experimentation of the Centro Nacional de Biotecnología (CNB) (Madrid, Spain) and by the Division of Animal Protection of the Comunidad de Madrid (PROEX, nos. 169.4/20 and 161.5/20). All animal procedures were performed according to the European Directive 2010/63/EU and the Spanish directive RD/53/2013 for the protection of animals used for scientific purposes. Sample sizes were determined on the basis of those reported in previous publications of our group30,31. Mice were allocated in the following experimental groups: controls (uninfected); SARS-CoV-2, 2-, 4- and 6 dpi; MVA-WT; MVA-S, 1 and 2 doses. Researchers were blinded to the details of the experimental groups during data collection and analysis.
Viruses
The poxviruses used in this study included the attenuated MVA-WT strain obtained from the Chorioallantois vaccinia virus Ankara strain after 586 serial passages in chicken embryo fibroblasts51 and the MVA-CoV2-S vaccine candidate expressing a human codon-optimized full-length SARS-CoV-2 S protein30.
The SARS-CoV-2 strain MAD6 (kindly provided by J. M. Honrubia and L. Enjuanes, CNB-CSIC) is a virus collected from a nasopharyngeal swab from a 69-year-old male patient with COVID-19 from Hospital 12 de Octubre, Madrid, Spain52. The growth and titration of SARS-CoV-2 MAD6 isolate have been described previously30,31. The full-length virus genome was sequenced and was identical to the SARS-CoV-2 reference sequence (Wuhan-Hu-1 isolate, GenBank no.: MN908947), except for the silent mutation C3037>T and two mutations leading to amino acid changes: C14408>T (in nsp12) and A23403>G (D614G in the S protein).
MVA-S vaccination and SARS-CoV-2 infection in K18-hACE2 mice
For experiments analyzing SARS-CoV-2 brain infection and neuropathological damage, female and male K18-hACE2 mice (4–5 months old; n = 26) were infected with SARS-CoV-2 (MAD6 strain, 1 × 105 PFU in 50 μl of PBS, intranasally) as described previously30,31.Uninfected control mice only received 50 μl of PBS by intranasal route (n = 9; 4 females and 5 males). Mice were euthanized at 2 (n = 8; 3 females and 5 males), 4 (n = 8; 3 females and 5 males) and 6 (n = 10; 5 females and 5 males) dpi; brains were extracted and fixed in 4% paraformaldehyde (PFA, Sigma-Aldrich) in PBS for at least 7 d.
MVA-CoV2-S immunization studies were carried out as indicated previously30,31. Briefly, in the experiments of a single SARS-CoV-2 infection, female K18-hACE2 mice (10 weeks old; n = 11 per group) received 1 or 2 doses of 1 × 107 PFU of MVA-CoV2-S in 100 μl of PBS (injected intramuscularly; 50 μl per leg) at 0 and 4 weeks. Also, mice were primed and boosted with nonrecombinant MVA-WT were used as the control group. At week 9, mice were challenged with SARS-CoV-2 as specified above. For the reinfection experiments, MVA-S-vaccinated mice were additionally reinfected with SARS-CoV-2 (MAD6 strain; 1 × 105 PFU in 50 μl of PBS; administered intranasally) 7 weeks after the first viral infection31. In this second set of experiments, mice treated with nonrecombinant MVA-WT and nonvaccinated, SARS-CoV-2-infected mice were used as controls. Mice were euthanized at 4 and 6 dpi, for single SARS-CoV-2 infection (MVA-WT: n = 9, 3 females and 6 males; MVA-S 1 dose: n = 9, 3 females and 6 males; MVA-S 2 doses: n = 3 females) and for the reinfection experiments (MVA-WT: n = 5 females; SARS-CoV-2: n = 4 females; MVA-S 1 dose: n = 5 females; MVA-S 2 doses: n = 4 females), respectively. Subsequently, the brains were extracted and fixed in 4% PFA for a period longer than 7 d.
Histological staining
Brains were cryoprotected in 30% sucrose (Sigma-Aldrich) in PBS and included in optimum cutting temperature compound (Tissue-Tek). Coronal sections (thickness 40 μm) were cut on a cryostat (Leica). SARS-CoV-2 N protein, NeuN, c-casp3, GFAP, IBA1, IB4, CaMKII, ChAT, DARPP32, oligodendrocytes (clone NS-1), tyrosine hydroxylase and parvalbumin immunohistological detection was performed as described previously41,53,54,55 using, respectively, mouse monoclonal anti-SARS-CoV-2, clone B46F (1:100 dilution, catalog no. MA1-7404, RRID: AB_1018422, Invitrogen); anti-NeuN (rabbit polyclonal, 1:500 dilution, catalog no. ABN78, RRID: AB_10807945, Millipore and mouse monoclonal, clone A60, 1:200 dilution, catalog no. MAB377, RRID: AB_2298772, Millipore); rabbit polyclonal anti-c-casp3 (1:100 dilution, catalog no. 9661, RRID: AB_2341188, Cell Signaling Technology); polyclonal anti-GFAP (rabbit polyclonal, 1:500 dilution, catalog no. Z0334, RRID: AB_10013382, Dako and mouse monoclonal, clone G-A-5, 1:2,000 dilution, catalog no. G3893, RRID: AB_477010, Sigma-Aldrich); anti-IBA1 (rabbit polyclonal 1:500 dilution, cat, no. 019-19741, RRID: AB_839504, Wako Chemicals and rabbit polyclonal, 1:1,000, catalog no. 234003, RRID: AB_10641962, Synaptic System); anti-IB4 (biotinylated isolectin B4, 1:50 dilution, catalog no. L2140, RRID: AB_2313663, Sigma-Aldrich); anti-CaMKII alpha/beta/delta (rabbit polyclonal, 1:100 dilution, catalog no. PA5-38239, RRID: AB_2554841, Thermo Fisher Scientific); anti-ChAT (chicken polyclonal, 1:500 dilution, catalog no. G143, Applied Biological Materials); anti-DARPP32 (rabbit polyclonal, 1:250 dilution, catalog no. PA5-85787, RRID: AB_2792923, Thermo Fisher Scientific); anti-oligodendrocytes, clone NS-1 (mouse monoclonal, 1:1,000 dilution, catalog no. MAB1580, RRID: AB_94266, Merck Millipore); anti-tyrosine hydroxylase (chicken polyclonal, 1:1,000 dilution, catalog no. TYH, RRID: AB_10013440, Aves Labs); anti-parvalbumin (rabbit polyclonal, 1:5,000 dilution, catalog no. PV27, RRID: AB_2631173, Swant); and secondary peroxidase-conjugated antibody kits (catalog nos. NB-23-00029-1 and NB-23-00030-1, NeoBiotech) or fluorescence secondary antibodies (goat-anti-mouse Alexa Fluor 488, 1:400 dilution, catalog no. 115-545-003, Jackson ImmunoResearch; goat-anti-rabbit Alexa Fluor 647, 1:400 dilution, catalog no. 111-605-003, Jackson ImmunoResearch; goat-anti-rabbit Alexa Fluor 568, 1:400 dilution, catalog no. A-11011, Invitrogen; streptoavidin-Cy3, 1:500 dilution, catalog no. 016-160-084, Jackson ImmunoResearch). In the case of SARS-CoV-2 and c-casp3 stained brain sections were subjected to citrate antigen retrieval (sodium citrate 10 mM, pH 6.0, Sigma-Aldrich; 15 min 97 °C); for SARS-CoV-2 immunodetection, brain sections were treated with mouse-on-mouse blocking reagent (catalog no. MKB2213-1, Vector Laboratories). In the immunofluorescence experiments, nuclei were stained with DAPI (1:1,000 dilution, Sigma-Aldrich). Nissl staining was performed as described previously56.
Image analysis and stereology
Image acquisition and analysis were performed with light transmission (Olympus, AX70 or Bx61, both with digital refrigerated camera DP72, CellSens v 1.4.1) or confocal microscopes (Nikon A1R+ or Leica STELLARIS 8 Scan Head, respectively, NIS-Element AR v 4.30.02 and LAS X v 4.3.024308) and their specific imaging software. Qualitative analysis of SARS-CoV-2 infection was performed by two independent blinded researchers. Imaging analyses of c-casp3, IBA1, GFAP, oligodendrocytes and IB4 were carried out as indicated previously41,54,55 using Fiji v 2.3.0 (National Institutes of Health) or Imaris microscopy image analysis software (Imaris, ×64 v.9.6.0, Oxford Instruments). NeuN+ and Nissl+ neuronal density was estimated by systematic random sampling using the optical dissector method57. Briefly, reference volumes were outlined at low magnification (×4) and neurons were counted at high magnification (×40) using a 4,900 × 30 μm2 optical dissector with a guard volume of 5 μm to avoid artifacts on the cut surface of the sections. All stereological procedures were performed using the New CAST system (Visiopharm) as described previously53,56. The confocal microglia images were analyzed in the Imaris software (×64 v.9.6.0). The microglial area and processes were measured using the Imaris surface and filament functions, respectively.
Analysis of SARS-CoV-2 RNA by RT–qPCR
The region corresponding to the olfactory bulb (bregma: +3.92–3.08 mm), cingulate cortex (bregma: +1.42 to −0.10 mm), hypothalamus (bregma: −1.82 to −2.18 mm) and brain stem (bregma: −5.50 to −7.08 mm) were microdissected from 3–6 coronal histological sections (thickness 40 μm) under a stereoscopic binocular microscope (Olympus SZX16) according to the mouse brain stereotaxic atlas58. RNA was isolated using the RecoverAll Total Nucleic Acid Isolation Kit (catalog no. AM1975, Invitrogen) according to the manufacturer’s instructions. The concentration and purity of the total RNA samples were measured using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). RNA integrity was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 LabChip kit (catalog no. 5067-1511; Agilent Technologies). For complementary DNA synthesis, 1 μg of RNA was reverse-transcribed with the QuantiTect Reverse Transcription Kit (catalog no. 205311, QIAGEN), according to the manufacturer’s specifications.
SARS-CoV-2 viral RNA content was determined using a previously validated set of primers and probes specific for the SARS-CoV-2 subgenomic RNA for the protein E59 and cellular 18S ribosomal RNA for normalization (catalog no. 4333760F, Thermo Fisher Scientific). Data were acquired with a 7500 real-time PCR system and analyzed with the 7500 software v.2.0.6 (Applied Biosystems). Relative RNA arbitrary units (a.u.) were quantified relative to the negative group (uninfected K18-hACE2 mice) and were performed using the \(2^{-{\Delta\Delta}_{\mathrm{Ct}}}\) method. All samples were tested in triplicate.
Statistical analysis
The number of mice analyzed in each experimental group and the statistical tests applied are indicated in each figure legend. Data are presented as the mean ± s.e.m. In all cases, normality and equal variance tests were performed; when passed, analysis of variance (ANOVA) with Dunnett, Tukey, Friedman or Fisher’s least significant difference (LSD) post hoc analysis for multiple groups, or unpaired t-tests for two-group comparisons, was carried out. In cases where normality or homoscedasticity tests failed, the nonparametric Kruskal–Wallis H test with post hoc Dunn’s test was performed. All statistical analyses were conducted using Prism v.8.0 (GraphPad Software).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All relevant data are included in the paper. This study did not generate datasets deposited in external repositories. Source data are provided with this paper.
Code availability
No custom code was used in this study.
References
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
Gupta, A. et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 26, 1017–1032 (2020).
Song, E. et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 218, e20202135 (2021).
Kumari, P. et al. Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. Viruses 13, 132 (2021).
Ramani, A. et al. SARS‐CoV‐2 targets neurons of 3D human brain organoids. EMBO J. 39, e106230 (2020).
de Melo, G. D. et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. 13, eabf8396 (2021).
Arbour, N., Day, R., Newcombe, J. & Talbot, P. J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 74, 8913–8921 (2000).
Netland, J., Meyerholz, D. K., Moore, S., Cassell, M. & Perlman, S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 82, 7264–7275 (2008).
Lewis, A. et al. Cerebrospinal fluid in COVID-19: a systematic review of the literature. J. Neurol. Sci. 421, 117316 (2021).
Matschke, J. et al. Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. 19, 919–929 (2020).
Winkler, E. S. et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020).
Oladunni, F. S. et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 11, 6122 (2020).
Zhou, B. et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592, 122–127 (2021).
Schlottau, K. et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe 1, e218–e225 (2020).
Jiao, L. et al. The olfactory route is a potential way for SARS-CoV-2 to invade the central nervous system of rhesus monkeys. Signal Transduct. Target. Ther. 6, 169 (2021).
Meinhardt, J. et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 24, 168–175 (2021).
Bauer, L. et al. The neuroinvasiveness, neurotropism, and neurovirulence of SARS-CoV-2. Trends Neurosci. 45, 358–368 (2022).
Yang, A. C. et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 595, 565–571 (2021).
Wenzel, J. et al. The SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat. Neurosci. 24, 1522–1533 (2021).
Schwabenland, M. et al. Deep spatial profiling of human COVID-19 brains reveals neuroinflammation with distinct microanatomical microglia-T-cell interactions. Immunity 54, 1594–1610 (2021).
Kanberg, N. et al. Neurochemical evidence of astrocytic and neuronal injury commonly found in COVID-19. Neurology 95, e1754–e1759 (2020).
Nalbandian, A. et al. Post-acute COVID-19 syndrome. Nat. Med. 27, 601–615 (2021).
Heneka, M. T., Golenbock, D., Latz, E., Morgan, D. & Brown, R. Immediate and long-term consequences of COVID-19 infections for the development of neurological disease. Alzheimers Res. Ther. 12, 69 (2020).
Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
Voysey, M. et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397, 99–111 (2021).
Sadoff, J. et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 384, 2187–2201 (2021).
Logunov, D. Y. et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet 396, 887–897 (2020).
Al Kaabi, N. et al. Effect of 2 Inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: a randomized clinical trial. JAMA 326, 35–45 (2021).
García-Arriaza, J. et al. COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike protein induce robust T- and B-cell immune responses and full efficacy in mice. J. Virol. 95, e02260-20 (2021).
Lázaro-Frías, A. et al. Full efficacy and long-term immunogenicity induced by the SARS-CoV-2 vaccine candidate MVA-CoV2-S in mice. NPJ Vaccines 7, 17 (2022).
Pérez, P. et al. A single dose of an MVA vaccine expressing a prefusion-stabilized SARS-CoV-2 spike protein neutralizes variants of concern and protects mice from a lethal SARS-CoV-2 infection. Front. Immunol. 12, 824728 (2022).
Boudewijns, R. et al. MVA-CoV2-S vaccine candidate neutralizes distinct variants of concern and protects against SARS-CoV-2 infection in hamsters. Front. Immunol. 13, 845969 (2022).
Mooij, P. et al. Poxvirus MVA expressing SARS-CoV-2 S protein induces robust immunity and protects rhesus macaques from SARS-CoV-2. Front. Immunol. 13, 845887 (2022).
Zheng, J. et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607 (2021).
Seehusen, F. et al. Neuroinvasion and neurotropism by SARS-CoV-2 variants in the K18-hACE2 mouse. Viruses 14, 1020 (2022).
Rothan, H. A. et al. SARS-CoV-2 infects primary neurons from human ACE2 expressing mice and upregulates genes involved in the inflammatory and necroptotic pathways. Pathogens 11, 257 (2022).
Olivarria, G. M. et al. Microglia do not restrict SARS-CoV-2 replication following infection of the central nervous system of K18-human ACE2 transgenic mice. J. Virol. 96, e0196921 (2022).
Haddad-Tóvolli, R., Dragano, N. R. V., Ramalho, A. F. S. & Velloso, L. A. Development and function of the blood–brain barrier in the context of metabolic control. Front. Neurosci. 11, 224 (2017).
Heine, V. M., Maslam, S., Zareno, J., Joëls, M. & Lucassen, P. J. Suppressed proliferation and apoptotic changes in the rat dentate gyrus after acute and chronic stress are reversible. Eur. J. Neurosci. 19, 131–144 (2004).
Alvarez-Vergara, M. I. et al. Non-productive angiogenesis disassembles Aß plaque-associated blood vessels. Nat. Commun. 12, 3098 (2021).
Bettcher, B. M., Tansey, M. G., Dorothée, G. & Heneka, M. T. Peripheral and central immune system crosstalk in Alzheimer disease—a research prospectus. Nat. Rev. Neurol. 17, 689–701 (2021).
McCray, P. B. Jr et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81, 813–821 (2007).
Khan, M. et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell 184, 5932–5949 (2021).
Krasemann, S. et al. The blood–brain barrier is dysregulated in COVID-19 and serves as a CNS entry route for SARS-CoV-2. Stem Cell Rep. 17, 307–320 (2022).
Halfmann, P. J. et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603, 687–692 (2022).
Dangi, T., Class, J., Palacio, N., Richner, J. M. & Penaloza MacMaster, P. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep. 36, 109664 (2021).
Ku, M. V. et al. Brain cross‐protection against SARS‐CoV‐2 variants by a lentiviral vaccine in new transgenic mice. EMBO Mol. Med. 13, e14459 (2021).
Taddeo, A. et al. Optimized intramuscular immunization with VSV-vectored spike protein triggers a superior immune response to SARS-CoV-2. NPJ Vaccines 7, 82 (2022).
Thepmankorn, P. et al. Cytokine storm induced by SARS-CoV-2 infection: the spectrum of its neurological manifestations. Cytokine 138, 155404 (2021).
Mayr, A., Stickl, H., Müller, H. K., Danner, K. & Singer, H. [The smallpox vaccination strain MVA: marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defense mechanism (author’s transl)]. Zentralbl. Bakteriol. B 167, 375–390 (1978).
Díez, J. M. et al. Cross-neutralization activity against SARS-CoV-2 is present in currently available intravenous immunoglobulins. Immunotherapy 12, 1247–1255 (2020).
Villadiego, J. et al. Long-term immunosuppression for CNS mouse xenotransplantation: effects on nigrostriatal neurodegeneration and neuroprotective carotid body cell therapy. Xenotransplantation 25, e12410 (2018).
Cabello-Rivera, D., Sarmiento-Soto, H., López-Barneo, J. & Muñoz-Cabello, A. M. Mitochondrial complex I function is essential for neural stem/progenitor cells proliferation and differentiation. Front. Neurosci. 13, 664 (2019).
March-Diaz, R. et al. Hypoxia compromises the mitochondrial metabolism of Alzheimer’s disease microglia via HIF1. Nat. Aging 1, 385–399 (2021).
Muñoz-Manchado, A. B. et al. Chronic and progressive Parkinson’s disease MPTP model in adult and aged mice. J. Neurochem. 136, 373–387 (2016).
West, M. J. New stereological methods for counting neurons. Neurobiol. Aging 14, 275–285 (1993).
Paxinos, G. & Franklin, K. B. J. The Mouse Brain in Stereotaxic Coordinates (Gulf Professional Publishing, 2004).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT–PCR. Euro Surveill. 25, 2000045 (2020).
Acknowledgements
We thank the CISA-INIA-CSIC for the biosafety level 3 facilities. The SARS-CoV-2 MAD6 virus isolate was kindly provided by J. M. Honrubia and L. Enjuanes (CNB-CSIC). We also thank to K. L. Levitsky and N. Borjini for technical assistance with the confocal acquisition and the Imaris software. We thank the CSIC and the Spanish Ministry of Science and Innovation for continuous support. This research was supported by the Spanish Ministry of Science and Innovation/Spanish Research Agency/10.13039/501100011033 grant nos. PID2019-105995RB-I00 (J.J.T.-A. and J.V.), PID2020-114481RB-I00 (J.G.-A. and M.E.), RTI2018-096629-B-I00 (A.P.) and PID2019-106410RB-I00 (J.L.-B.). Moreover, this research was also funded by Red TerCel ISCIII (no. RD16/0011/0025 to J.J.T.-A.), Consejería de Economía, Conocimiento, Empresas y Universidad US-1380891 (to J.J.T.-A. and J.V.), Consejería de Salud y Familias, Junta de Andalucía grant no. PECOVID-0078-2020 (to R.R.-L. and J.V.), Consejería de Educación y Deporte, Junta de Andalucía grant no. PY20_01312 (to A.P.), Fondo COVID-19 grant no. COV20/00151 (Spanish Health Ministry, Instituto de Salud Carlos III), Fondo Supera COVID-19 (Crue Universidades-Banco Santander) grant and CSIC grant no. 202120E079 (J.G.-A.), CSIC grant no. 2020E84, La CaixaImpulse grant no. CF01-00008 and Ferrovial and MAPFRE donations (to M.E.). Additionally, we received funding from the European Commission-NextGenerationEU, through the CSIC’s Global Health Platform (PTI Salud Global) (to J.G.-A. and M.E.) and the European Research Council (ERC Advanced grant no. PRJ201502629) (to J.L.-B.). J.G.-A. and M.E. also acknowledge financial support from the Spanish State Research Agency (no. AEI/10.13039/501100011033) through the ‘Severo Ochoa’ Programme for Centres of Excellence in R&D (nos. SEV-2013-0347 and SEV-2017-0712). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.V., J.G.-A., A.P., M.E., J.L.-B. and J.J.T.-A. conceptualized the study. J.V., J.G.-A., R.R.-L., A.P., M.E., J.L.-B. and J.J.T.-A. acquired the funding. J.V., J.G.-A., R.R.-L., R.G.-S., D.C.-R., A.E.R.-N., M.I.A.-V., F.C.-F., E.G.-R., J.L.L.-O., C.Z., D.A., G.A., P.P., A.M.M.-C. and A.P. devised the methodology and carried out the investigation. J.V., A.M.M.-C. and A.P. analyzed the data. J.V., J.G.-A., A.P., M.E., J.L.-B. and J.J.T.-A. supervised the study. J.V., A.M.M.-C. and A.P. visualized the data. J.V. and J.J.T.-A. wrote the original draft. J.V., J.G.-A., R.G.-S., A.M.M.-C., A.P., M.E., J.L.-B. and J.J.T.-A. reviewed the draft. All authors read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Michael Heneka, Stanley Perlman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of SARS-CoV-2 infection in the principal brain regions of K18-hACE2 mice.
a, High magnification images, after SARS-CoV-2 N immunohistochemistry, from control and SARS-CoV-2 infected K18-hACE2 mice (at 2, 4 and 6 dpi) of the indicated cerebral regions showing the level of SARS-CoV-2 infection. b, Quantitative analysis of SARS-CoV-2 RNA at 2 dpi, detected by RT-qPCR targeting the viral E gene, in the olfactory bulb, cortex, hypothalamus and brainstem. Data are presented as mean ± standard error of the mean (SEM). Uninfected control mice, n = 5 (3 females and 2 males); SARS-CoV-2 infected mice at 2 dpi, n = 8 (3 females and 5 males). The data of SARS-CoV-2 infected mice at 2 dpi are also represented in Fig. 1d, but with different scale.
Extended Data Fig. 2 Analysis of SARS-CoV-2 infection in the hippocampus.
a, High magnification images, after SARS-CoV-2 N immunohistochemistry, from control and SARS-CoV-2 infected K18-hACE2 mice (at 2, 4 and 6 dpi) showing the viral infection in CA1, CA2/CA3 and dentate gyrus. b, Qualitative analysis of SARS-CoV-2 level of infection in the hippocampal regions described above of infected K18-hACE2 mice at 2, 4, and 6 dpi. Between brackets the number of mice showing SARS-CoV-2+ cells among the total number of mice studied is indicated for each region. c, Stereological estimation of NeuN+ neurons in the indicated hippocampal regions. Data are presented as mean ± standard error of the mean (SEM). Uninfected controls, n = 9 (4 females and 5 males); SARS-CoV-2 infected mice at 6 dpi, n = 10 (5 females and 5 males). Unpaired, two-sided, t-test, dentate gyrus: p = 0.0037. **p < 0.05, respect control.
Extended Data Fig. 3 Analysis of SARS-CoV-2 infection in specific neuronal subtypes.
Microscopy confocal images showing the colocalization of SARS-CoV-2 (green) with neuronal subtype specific markers (in red): parvalbumin (PV; a); tyrosine hydroxylase (TH, b); and DARPP32 (c) showing the viral infection of striatal GABAergic-PV+ neurons, hypothalamic dopaminergic-TH+ neurons and striatal DARPP32+ medium spiny neurons, respectively. The insets are depicted, at higher magnification, on the right. Arrows label some examples of SARS-CoV-2 infected neurons. Nuclei were counterstained with DAPI (blue). The immunofluorescence stainings were performed in 3 independent experiments obtaining similar results, analyzing SARS-CoV-2 infected mice at 6 dpi, n = 6 (3 females and 3 males). In b, 3 V: third ventricle.
Extended Data Fig. 4 Colocalization analysis of SARS-CoV-2 N protein in glial or vascular brain cells.
a-c, High-resolution Z-projection confocal images from the cortex of a SARS-CoV-2 infected K18-hACE2 mouse (6 dpi), after immunohistological detection of SARS-CoV-2 N protein (green and GFAP (red). The insets are depicted at higher magnification, in b and c, exhibiting single confocal planes (z = 14 and 3 in b; z = 4 in c) of 405 nm z-depth optic resolution and the orthogonal projections. Note the absence of colocalization between green (SARS-CoV-2 N protein) and red (GFAP) fluorescence signals. Arrows in c indicates an example of a GFAP+ process without SARS-CoV-2+ fluorescence signal. d, Cortical confocal images from a SARS-CoV-2 infected K18-hACE2 mouse (6 dpi) immunostained with SARS-CoV-2 N protein (green) and a vascular (IB4; red) marker. Note the presence of a discrete dot of green fluorescence signal (SARS-CoV-2 N protein+) in vascular IB4+ cell, pointed by the arrow and showed by the YZ-orthogonal projection in the left (merge) panel. e, High-resolution Z-projection confocal images from the cortex of SARS-CoV-2 infected K18-hACE2 mouse (6 dpi) immunolabelled with SARS-CoV-2 N protein (green) and the microglial marker IBA1 (red), the inset is showed at higher magnification in h. f,g. High resolution confocal image and 3D reconstructions showing the phagocytic activity of IBA1+ cells on a SARS-CoV-2 infected neuron (f) or brain vascular cells marked by arrows in g. h. High resolution confocal image of the region marked by the inset in e. Arrows and insets show the presence of discrete dots of green fluorescence signal (SARS-CoV-2 N protein+) inside of a microglial cell. These are exhibited at higher magnification in h’ and h’ as single confocal planes (z = 3 and z = 17, respectively; 401 nm z-depth optic resolution) and orthogonal projections. The immunofluorescence stainings were performed in 3 independent experiments obtaining similar results, analyzing SARS-CoV-2 infected mice at 6 dpi, n = 6 (3 females and 3 males).
Extended Data Fig. 5 SARS-CoV-2 cerebral infection produces cellular apoptosis.
a, Light-transmitted images of the indicated cerebral regions from control and SARS-CoV-2 infected K18-hACE2 mice (6 dpi) after c-caspase-3 immunohistochemistry. Images in the right column depict, at higher magnification, the insets of the central column. c-caspase-3 immunostainings were performed in 4 independent experiments obtaining similar results. b, Qualitative analysis of cellular apoptosis, induced by SARS-CoV-2 infection, in different cerebral regions. The table indicates the number of mice showing c-casp3+ cells above the level in control mice among the total number of mice studied for each brain region analyzed.
Extended Data Fig. 6 Glial response and vascular damage in SARS-CoV-2 infected brain.
Confocal Z-projection images from control and SARS-CoV-2 infected K18-hACE2 mice stained with microglial (IBA1; cyan), vascular (IB4; red), and astrocytic (GFAP, green) markers. The insets are depicted at higher magnification in Fig. 4h. The immunofluorescence stainings were performed in 5 independent experiments obtaining similar results.
Extended Data Fig. 7 MVA-CoV2-S vaccination prevents SARS-CoV-2 brain infection.
Images of light-transmitted microscopy, after SARS-CoV-2 N immunohistochemistry, of the indicated cerebral regions showing the total protection conferred by the MVA-S vaccine (1 or 2 doses) against cerebral SARS-CoV-2 infection, in the reinfection experiment (6 dpi). SARS-CoV-2 N immunostainings were performed in 5 independent experiments obtaining similar results.
Extended Data Fig. 8 Histological analysis of glial cells and brain vasculature after MVA-CoV2-S vaccination.
Representative confocal Z-projection images from the hypothalamus of mice subjected to the SARS-CoV-2 reinfection experiment (controls; SARS-CoV-2; MVA-S 1 dose; MVA-S 2 doses) which were stained with microglial (IBA1; cyan), and vascular (IB4; red) markers. Nuclei were counterstained with DAPI (blue) in merge images. The immunofluorescence stainings were performed in 4 independent experiments obtaining similar results.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Villadiego, J., García-Arriaza, J., Ramírez-Lorca, R. et al. Full protection from SARS-CoV-2 brain infection and damage in susceptible transgenic mice conferred by MVA-CoV2-S vaccine candidate. Nat Neurosci 26, 226–238 (2023). https://doi.org/10.1038/s41593-022-01242-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01242-y